The world’s simplest known animal is very poorly understood
World’s Simplest Animal Reveals Hidden Diversity
The first animal genus defined purely by genetic characters represents a new era for the sorting and naming of animals
· By Charlie Wood, Quanta Magazine on October 6, 2018
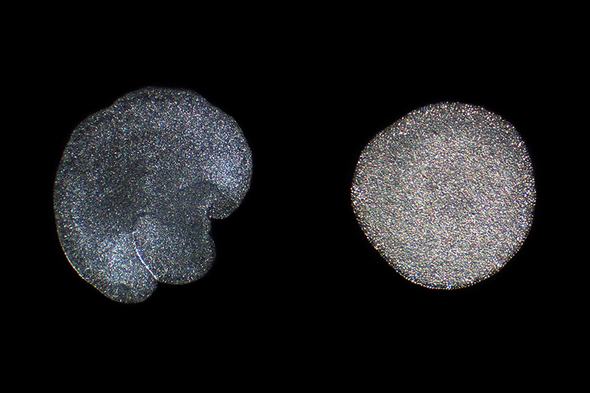
Two individuals
from two different genera of the phylum Placozoa: Trichoplax adhaerens (left) and Hoilungia hongkongensis (right). The gross morphology and the internal structure
are indistinguishable, and only genetics can tell them apart. Credit: Hans-Jürgen Osigus, ITZ Ecology and Evolution, Stiftung
Tierärztliche Hochschule Hannover, Germany
The world’s simplest known animal is so poorly understood that
it doesn’t even have a common name. Formally called Trichoplax adhaerens for the way it adheres to
glassware, the amorphous blob isn’t much to look at. At just a few millimeters
across, the creature resembles a squashed sandwich in which the top layer
protects, the bottom layer crawls, and the slimy stuffing sticks it all
together. With no organs and just a handful of cell types, the most
interesting thing about T. adhaerens might
just be how stunningly boring it is.
“I
was fascinated when I first heard about this thing because it has no real
defined body,” said Michael Eitel, an
evolutionary biologist at the Ludwig Maximilian University in Germany. “There’s
no mouth, there’s no back, no nerve cells, nothing.”
But
after spending four years painstakingly reconstructing the blob’s genome, Eitel
might know more about the organism than anyone else on the planet. In
particular, he has looked closely enough at its genetic code to learn what
visual inspections failed to reveal. The variety of creature that biologists
have long called T. adhaerens is really at
least two, and perhaps as many as a dozen, anatomically identical but genetically distinct “cryptic species” of
animals. The discovery sets a precedent for taxonomy, the
science of naming organisms, as the first time a new animal genus has been
defined not by appearance, but by pure genetics.
The
modern taxonomic system, little changed since Carl Linnaeus laid it out in the
1750s, attempts to chop the sprawling tree of life into seven tidy levels that
grant every species a unique label. The two-part scientific name (such as Homo sapiens) represents the tail end of a branching
path through this tree, starting from the thickest limbs, the kingdoms, and
ending at the finest twigs, the genus (Homo) and then the
species (sapiens). The path tells you everything there is to
know about the organism’s relationship to other groups of creatures, at least
in theory.
Ever
since its discovery in the late 1800s, T. adhaerenshas been
recognized as having a highly unusual body plan, and it has formally had the
phylum of Placozoa (“flat animals”) to itself for almost half a century. Just
one level more specific than kingdom, a phylum is a cavernous space to occupy
alone: Our phylum, Chordata, overflows with more than 65,000 living species
ranging from peacocks to whales to eels. Biologists have long suspected that
Placozoa hid more diversity, and mitochondrial evidence strengthened that
suspicion in 2004, when researchers found that short sequences from different
individuals looked about as different as those of organisms from different
families (one level more general than genus).
But
that observation about the two Placozoa didn’t meet the accepted international standards
for putting them in new taxonomic categories, which have historically been
based on animals’ forms. “At the time we had just uncovered the genetic
differences,” said Allen G. Collins, a
co-author of the 2004 paper and a zoologist at the National Systematics
Laboratory of the National Oceanic and Atmospheric Administration (NOAA).
“Looking at the animals we had collected, it wasn’t discernible how
they might differ morphologically.”
To
finish what Collins started, Eitel and his colleagues decided to abandon the
visual approach and search for defining characteristics in the placozoan genome
itself.
They
began by mapping out the phylum’s genetic territory with the same easy-to-sequence
mitochondrial DNA Collins had used. By comparing data from this molecule, known
as 16S, Eitel concluded that a particular variety of Placozoa from Hong Kong
was the most distant relative of the standard strain, the genome of which had
already been fully sequenced in 2008. If any group would qualify as a different
species, this was the one.
He
next needed to read, order and interpret the 80-odd million A, G, C and T
nucleotide bases that make up the Hong Kong variant’s genome. Growing a few
thousand placozoans, blending them to extract their nuclear DNA and converting
the snippets of their genome into a digital format took a few weeks, but the
hard work of shuffling those pieces into the right order and figuring out what
each section does took four years of fiddling around with computer programs.
When the team finally had a full genome ready for comparison, the payoff turned
out to be worth the wait. “We expected to find differences, but when I first
saw the results of our analyses, I was really overwhelmed,” Eitel said.
A quarter of the genes were in the wrong spot or written
backward. Instructions for similar proteins were spelled nearly 30 percent
differently on average, and in some cases as much as 80 percent. The Hong Kong
variety was missing 4 percent of its distant cousin’s genes and had its own
share of genes unique to itself. Overall, the Hong Kong placozoan genome was
about as different from that of T. adhaerens as human DNA is from mouse DNA. “It was
really striking,” Eitel said. “They look the same, and we look completely
different from mice.”
So
where do all those genetic changes manifest, if not in the animals’ flabby
appearance?
“Even
though the placozoan itself looks like a little ball of glue, it probably has
cells that are doing some pretty sophisticated things,” said Holly Bik, a marine biologist at the
University of California, Riverside, who studies tiny marine roundworms known
as nematodes, which can also be cryptic. The Hong Kong Placozoa came from a
brackish mangrove stream where large swings in temperature and salinity demand
flexible body chemistry. “Physiologically, for
organisms, that’s a pretty big thing to have to deal with. At the
molecular level you need specific adaptations,” said Bik, who was not involved
in the research.
By
comparing the Placozoa variation with the average genetic differences between groups
in other phyla, the German team concluded that the Hong Kong Placozoa
qualified as not only a new species, but also a new genus. It might even have
qualified as a new family or order in other areas of the animal tree, but
to err on the conservative side, the team based their standard of genus
variation on jellyfish, a genetically diverse phylum with relatively tidy
divisions between levels.
All
that remained was the naming. Taxonomic codes demand
identifying characteristics, but don’t specify whether they should be visual or
genetic, so the team picked out four genetic letters in the 16S mitochondrial
genome that could uniquely differentiate the two lineages. Then, endorsed by
peer review and PLOS Biology in late July,
their work placed a new organism on our map of life.
The
team gave their specimen the genus name Hoilungia, for a
shapeshifting dragon king from Chinese mythology, and they named the
species hongkongensis, for where it was collected. Similar
genome-based classifications are common in the protist and bacterial worlds, and a
relative handful of cryptic animal species have been named based on
genetics. Namings (and renamings) that blend morphological characters with
genetic ones, which recently re-classified a common
houseplantSusanne Renner, a
botanist at the Ludwig Maximilian University. “It’s just great.”
The
researchers hope their work will make it easier for future genetic
character–based naming, which is less subject to biases from attention-grabbing
visual features like antlers and fins that may not accurately reflect evolutionary
distance between groups. “Someone had to be the first one to fight for the
right to define new general species based on genomics, and we luckily got it
published,” Eitel said.
Renner
says this work is the latest step in an ongoing shift toward genetic taxonomy.
“It took a long time to take off and now it’s taking off,” she said. She points
out that in contrast with the pages of text that can go into a formal
description of a species, specifying an organism with just four letters as the
German team has done lends itself to snappy efficiency. “Linnaeus would be
happy to do it. He was envisioning very brief and sharp diagnoses.”
As
precise as genetic classification can be however, it will likely complement
traditional ways of telling animals apart, not replace them. Observing visual
features doesn’t require years of a lab’s time. Even for other cryptic animals
like nematodes, which can’t be raised in captivity, genetic techniques may find
limited use. “For me, working with a single nematode worm, there’s never going
to be enough DNA isolated from an individual to use some of these
technologies,” Bik said.
But
for cryptic animals that researchers can cultivate, genetic sequencing may be
the perfect spotlight for illuminating the shaded parts of their evolutionary
tree. Eitel said he learned a lot from the process of analyzing the H. hongkongensis genome and predicts that
sequencing the next variant—a project already underway—will take months, not
years. “There will probably be dozens of new species popping up in the future,”
he said. “And more to come, because we’re constantly sampling.”
Reprinted with permission from Quanta
Magazine, an editorially independent publication of
the Simons Foundation whose mission is to enhance public understanding of science
by covering research developments and trends in mathematics and the physical
and life sciences.
[[My friend Dovid Moshe Yasnyi offers a very useful snapshot of the history of classification:
[[My friend Dovid Moshe Yasnyi offers a very useful snapshot of the history of classification:
"The
history of taxonomy.
There
are three periods.
Era
one: From Aristotle to Darwin
Names
of living things reflected attributes, later focusing on form (morphology) and
function (physiology). Linnaeus systematized it so that the names of different
organisms contained information about the relationship between the organisms.
Era
Two: From Darwin to Watson and Crick
Names
based on evolutionary closeness.
Era
Three: After Watson and Crick
Names
based on genetic closeness which is understood to mean evolutionary closeness,
but in practice involves a different approach."]]